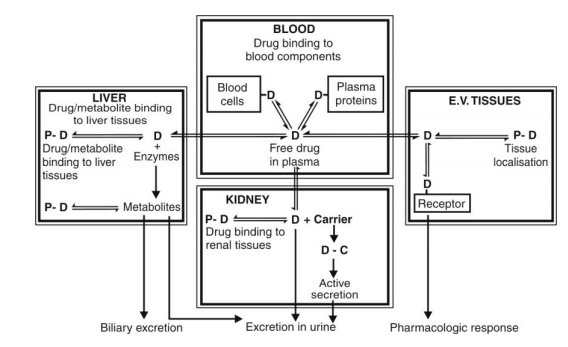
Uncovering Drug Mechanism of Action by Proteome Wide- Identification of Drug-Binding Proteins | Bentham Science

Binding mechanism of triclocarban with human serum albumin: Effect on the conformation and activity of the model transport protein - ScienceDirect

Identification of the Type I Collagen-binding Domain of Bone Sialoprotein and Characterization of the Mechanism of Interaction* - Journal of Biological Chemistry

a Schematic diagram of the MmfR binding mechanism. Binding of MmfR to... | Download Scientific Diagram
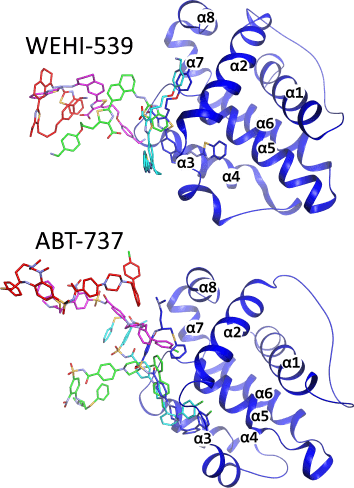
BSM00021: Cryptic-site binding mechanism of medium-sized Bcl-xL inhibiting compounds elucidated by McMD-based dynamic docking simulations - BSM-Arc

A schematic diagram describing the proposed two-step binding mechanism for proteins in steric occlusion of the direct binding of the ligands.

Universality of the Sodium Ion Binding Mechanism in Class A G‐Protein‐Coupled Receptors - Selvam - 2018 - Angewandte Chemie International Edition - Wiley Online Library

Unraveling the Binding Mechanism of Trivalent Tumor Necrosis Factor Ligands and Their Receptors* - Molecular & Cellular Proteomics

Insights into the substrate binding mechanism of SULT1A1 through molecular dynamics with excited normal modes simulations | Scientific Reports
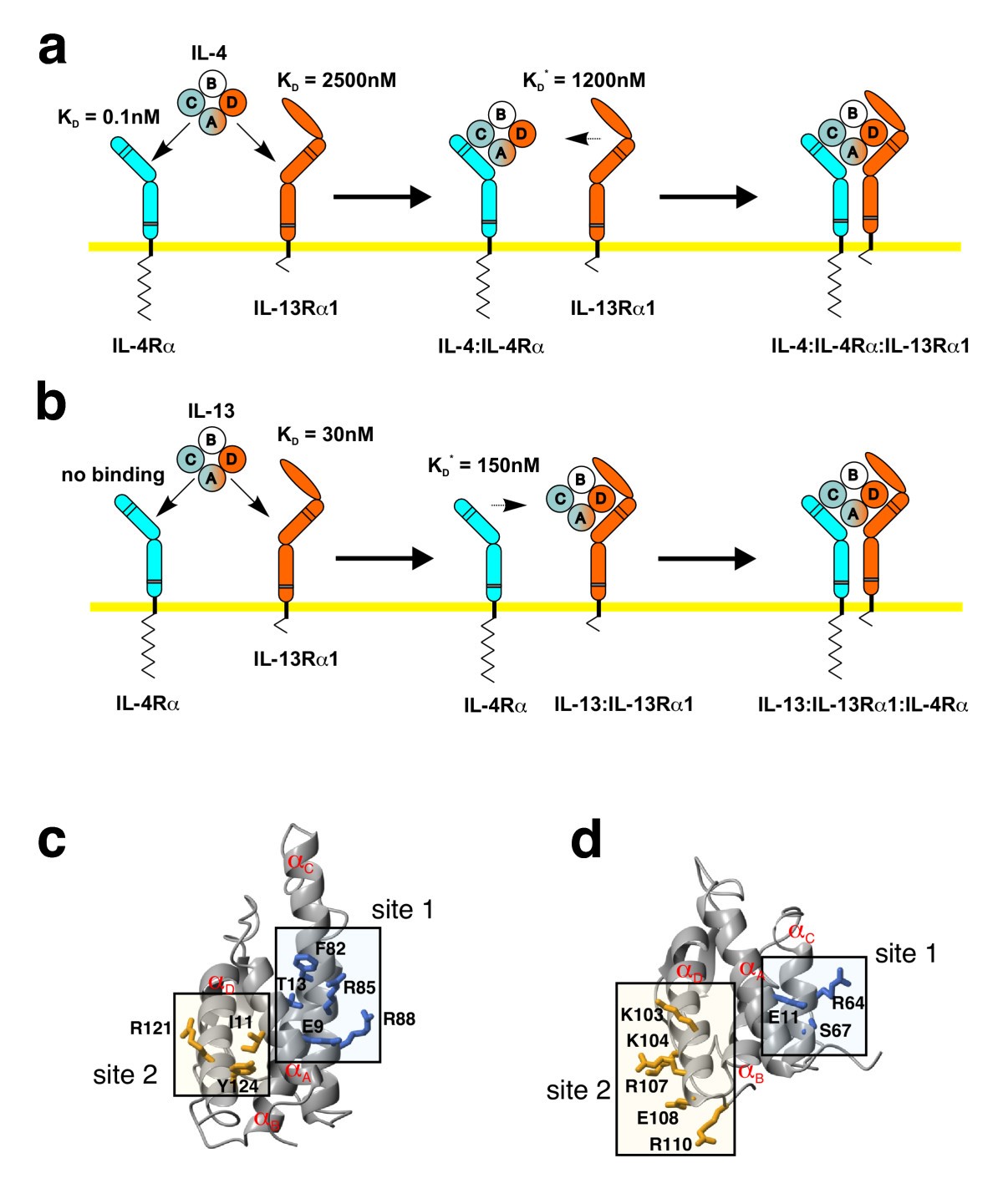
A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor | BMC Biology | Full Text

Binding mechanism and binding free energy of amino acids and citrate to hydroxyapatite surfaces as a function of crystallographic facet, pH, and electrolytes - ScienceDirect
PLOS Computational Biology: Lipidated apolipoprotein E4 structure and its receptor binding mechanism determined by a combined cross-linking coupled to mass spectrometry and molecular dynamics approach

Both protein dynamics and ligand concentration can shift the binding mechanism between conformational selection and induced fit | PNAS
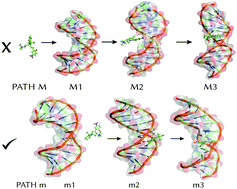
DNA-binding mechanism of spiropyran photoswitches: the role of electrostatics - Physical Chemistry Chemical Physics (RSC Publishing)